Weizmann Institute of Science-led researchers have unveiled a computational workflow that crafts enzymes exhibiting catalytic efficiencies surpassing 100,000 M−1 s−1, achieving performance comparable to natural biocatalysts without extensive refinement.
Natural enzymes perform catalytic biochemical transformations with extraordinary speed and precision, setting a benchmark that artificial catalysts have struggled to meet. Computational strategies have produced proteins capable of catalyzing reactions outside the repertoire of natural enzymes, with conversion rates that are often many orders of magnitude below the speeds recorded in biologically evolved catalyst reactions.
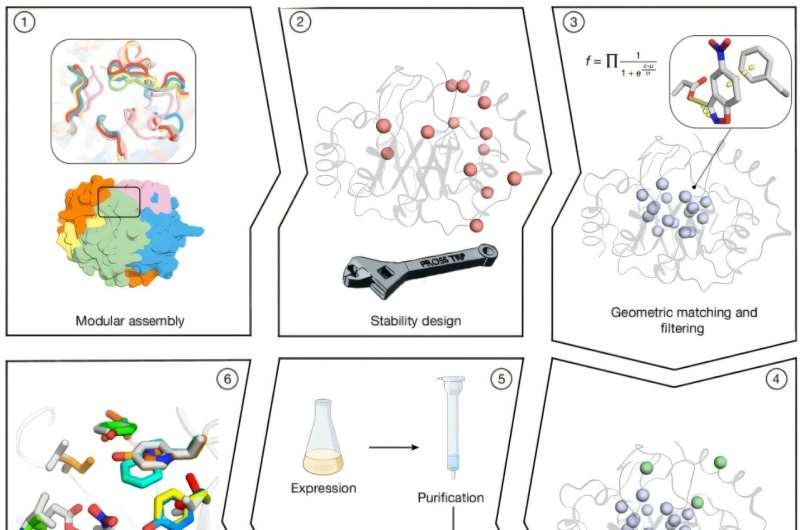
In the study, “Complete computational design of high-efficiency Kemp elimination enzymes,” published in Nature, researchers developed a fully computational approach to create stable, active enzymes with minimal optimization.
Researchers generated thousands of TIM-barrel backbones (one of the most prevalent protein folds found among enzymes) through combinatorial assembly and design, combining fragments from homologous proteins. In this fold, the residues of the central β barrel are oriented towards the active-site cavity, providing many opportunities for optimally placing the catalytic and substrate-binding groups.
Active-site residues were optimized using Rosetta atomistic calculations, yielding millions of designs that were filtered by an objective function balancing energy and desolvation of the catalytic base. A total of 73 designs were selected for experimental testing; 66 were solubly expressed, and 14 exhibited cooperative thermal denaturation behavior.
Introducing 5–8 active-site mutations per variant produced designs with increased catalytic efficiencies. One variant derived from Des61 (design 61) reached a kcat/KM of 3,600 M−1 s−1 (a measure of catalytic efficiency) and a turnover rate of 0.85 s−1 (reactions per second).
Eight designs based on Des27 achieved catalytic rates 10–70-fold higher than the original, with Des27.7 attaining a kcat/KM of 12,700 M−1 s−1 and kcat of 2.85 s−1. A single-point mutation replacing Phe113 with leucine raised catalytic efficiency to 123,000 M−1 s−1 and kcat to 30 s−1.
Researchers conclude that active-site preorganization combined with the capacity to adopt multiple catalytically competent substrate-bound conformations distinguishes this design from earlier examples. The best variant reached high stability (greater than 85 °C) and catalytic efficiency comparable to natural enzymes.
Results confirm that atomistic computational methods are already sufficiently reliable to produce efficient enzymes in natural folds without iterative laboratory screening or reliance on artificial intelligence-generated scaffolds.
Future advances in modeling theoretical enzymes may enable fully programmable biocatalysis capable of performing a wide array of untapped solutions, from product manufacturing and therapeutic applications to pollution remediation, green chemistry and industrial scale molecular synthesis.
Written for you by our author Justin Jackson,
edited by Sadie Harley, and fact-checked and reviewed by Andrew Zinin—this article is the result of careful human work. We rely on readers like you to keep independent science journalism alive.
If this reporting matters to you,
please consider a donation (especially monthly).
You’ll get an ad-free account as a thank-you.
More information:
Dina Listov et al, Complete computational design of high-efficiency Kemp elimination enzymes, Nature (2025). DOI: 10.1038/s41586-025-09136-2
Zhuofan Shen et al, Highly efficient enzymes designed from scratch, Nature (2025). DOI: 10.1038/d41586-025-02054-3
© 2025 Science X Network
Citation:
Scientists design stable enzymes for non-natural reactions with near-natural efficiency (2025, July 14)
retrieved 14 July 2025
from https://phys.org/news/2025-07-scientists-stable-enzymes-natural-reactions.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.